IN A NUTSHELL
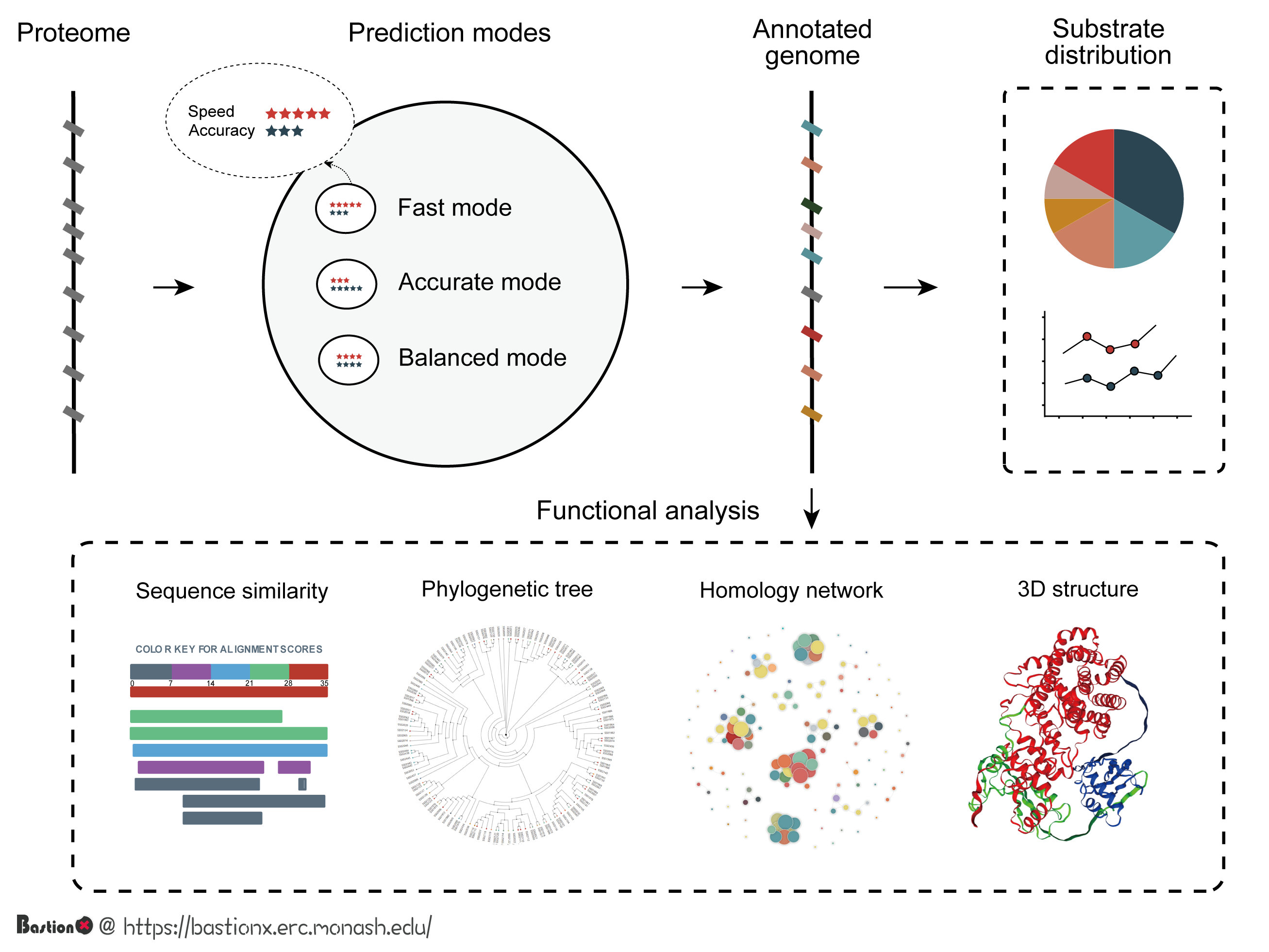
1. About This Work
Gram-negative bacteria have evolved an extraordinary array of secretion systems to export protein substrates into target cells or the surrounding environment. These protein substrates differ significantly in their structures and functions, and in the dedicated secretory pathway that they use. Accordingly, it is difficult to develop computational systems for accurate prediction of substrate type.
In this work, we present an integrative system, BastionX, to predict and visualize various types of secreted substrates in Gram-negative bacteria. BastionX outperforms existing substrate predictors by four major upgrades: 1) BastionX is the first predictor of types 1 and 2 secreted substrates, and also includes more accurate predictions for types 3, 4, and 6 secretion systems; 2) integrating single predictors within a framework, BastionX can seamlessly annotate various types of secreted substrates in a given bacterial genome and provides interactive visualization for its general substrate distribution; 3) Specifically, for each predicted substrate, BastionX enables further functional analysis within four interactive visualizations; and 4) BastionX provides three modes to cater to different needs: Fast mode for rapid prediction, Accurate mode for accurate prediction, and Balanced mode for a balance between prediction accuracy and speed. In combination with its additional standalone toolkit, BastionX can be conveniently executed locally to conduct sequence analysis and be readily integrated into a user's own pipeline. Taken together, BastionX can simultaneously annotate thousands of protein sequences with their potential secretion substrate type and map the global landscape of how secreted substrates are distributed across bacterial genomes.
DATASETS
1. Construction of the Training Dataset and the Independent Test Dataset
For each type of secreted substrate, we conducted an exhaustive and thorough literature search to construct the benchmark dataset, followed by a redundancy reduction procedure using the CD-HIT (Huang, et al., 2010) at a cutoff threshold of 0.7. Accordingly, we obtained 161, 79, 504, 414 and 148 protein sequences for type I, II, III, IV and VI substrates, respectively. The curated data for each type of substrate was randomly split into the training dataset (80%) and independent dataset (20%) as positive samples. For each substrate type, 1112 non-substrates used in previous work ((Wang, et al., 2019; Wang, et al., 2019; Wang, et al., 2018; Zou, et al., 2013)) were included as negative samples in the training dataset. Non-substrates retrieved from the UniProt database were included as negative samples to construct the independent dataset with a ratio of 1:1 between positive and negative samples.
2. Case Study Sequences
We further selected five substrates as a case study to investigate the superior prediction performance of BastionX, as compared with single type predictors. These substrates are associated with different secretion systems, but each of them contain common shared characteristics that might be incorrectly recognized by more than one single type predictors.
ONLINE WEB SERVER
1. BastionX
To maximize users' convenience, we have developed a user-friendly and easy-to-use web server, termed BastionX, to provide public service to the scientific community.
2. Using the BastionX Web Server
BastionX is an online server implemented with a user-friendly interface, which makes it very easy to use. It allows users to submit the interested sequences in the input page.
2.1 Input Formats
Two types of input are accepted by BastionX: sequences in FASTA format (strongly recommended) and raw sequences.
In the case of input sequences in the FASTA format, you can prepare and input them as follows:
In the case of raw sequences, you can input them as follows:
which will be formated by BastionX as follows:
2.2 Input Sequence Limits
1. The length of each submitted sequence should be in the range of 30 and 5000.
2. Considering that substrate prediction is a little bit time-consuming, the maximum number of sequences allowed for each submission by the BastionX server should be no more than 50000.
Note: These limitations don't apply to the BastionX Standlone Software
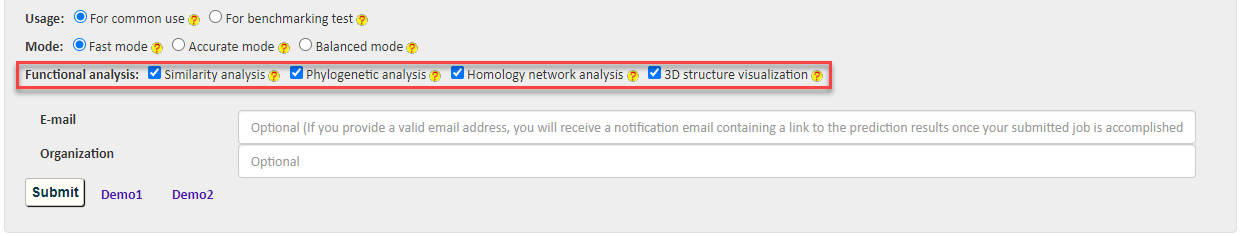
Besides, BastionX provides a range of options to accelerate computational prediction and enrich functional analyses. Users can select the following options to customize for their own purpose.
2.3 Usage
There are two options for users' choice in different scenarios: 1) For common use to provide service in practical scenario and 2) For benchmarking test to allow users to conduct benchmark test on the server.
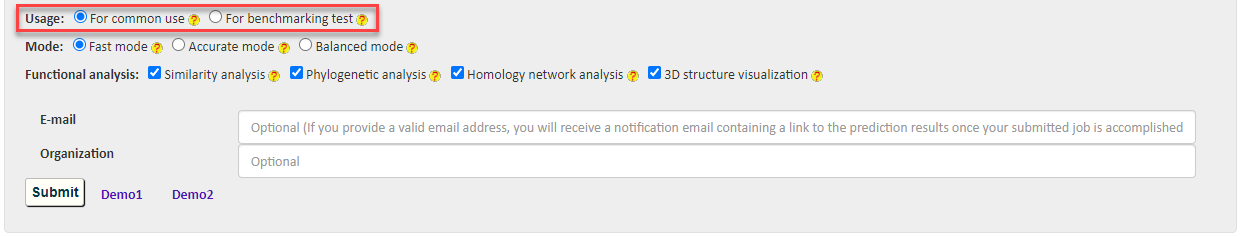
For common use: The BastionX server stores a built-in list of experimentally validated secreted substrates to filter the query proteins prior to model prediction. As a result, the query protein at the result page will be labeled as "Exp" with a score of 1 if it is a known, experimentally validated secreted protein (with a hit in the built-in list), and otherwise, labeled as "Pred" with a predicted score.
For benchmarking test: This option allows users to disable the built-in list of experimentally validated substrate proteins, to retrieve the prediction stores for all query proteins.
2.4 Mode
BastionX provides three modes to cater to different needs: 1) Fast mode for rapid prediction, 2) Accurate mode for accurate prediction, and 3) Balanced mode for a balance between prediction accuracy and speed.
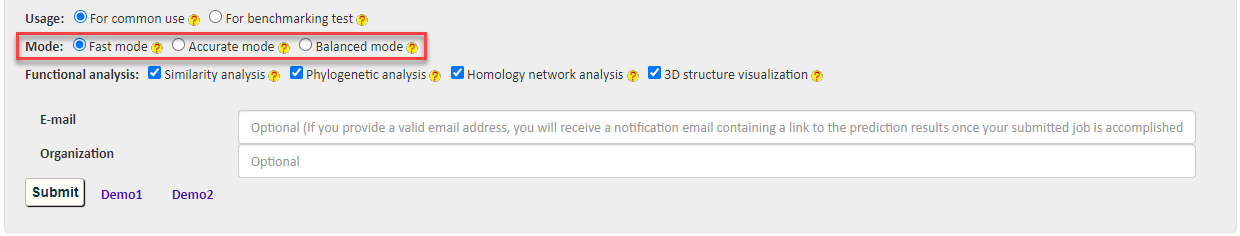
Fast mode : When Fast mode is activated, the BastionX server will invoke the 'fast-oriented' model that was constructed based on sequence-derived and physicochemical property-based features to generate the prediction results. This mode is suitable for preliminary screen of substrates in large-scale protein sequences.
Accurate mode: When Accurate mode is activated, the BastionX server will invoke the 'accurate-oriented' model that was constructed using the same methodology as the Fast mode, but based on evolutionary features (which is a well-demonstrated type of highly informative but time-consuming features). This mode is suitable for accurate identification of target proteins.
Balanced mode: When Balanced mode is activated, the BastionX server will first invoke the Fast mode model to do a preliminary screen, and then pass the possible substrates (using a low prediction threshold of 0.3 to reduce omission of real substrates) to the Accurate mode model for final prediction. This mode represents a balance between prediction accuracy and speed.
2.5 Functional Aanalysis
BastionX provides four modules to encourage users to make further functional analysis, including 1) Similarity analysis, 2) Phylogenetic analysis, 3) Homology network analysis and 4) interactive 3D structure visualization.
Similarity analysis: Similarity analysis finds regions of similarity between biological sequences. It uses blast 2.8.1+ to search inquiry protein sequences against experimentally validated substrates to calculates the statistical significance and visualizes the sequence similarities by BlasterJS.
Phylogenetic analysis: Phylogenetic analysis recognizes the closest relationships of inquiry proteins among experimentally validated substrates based on sequence multiple alignment using MAFFT v7.310 , and visualizes them in form of a phylogenetic tree by the open source phylogram_d3 .
Homology network analysis: Homology network analysis recognizes the closest relationships of inquiry proteins with experimentally validated substrates based on the all-against-all BLAST (version blast-2.2.26) and visualizes them by ECharts .
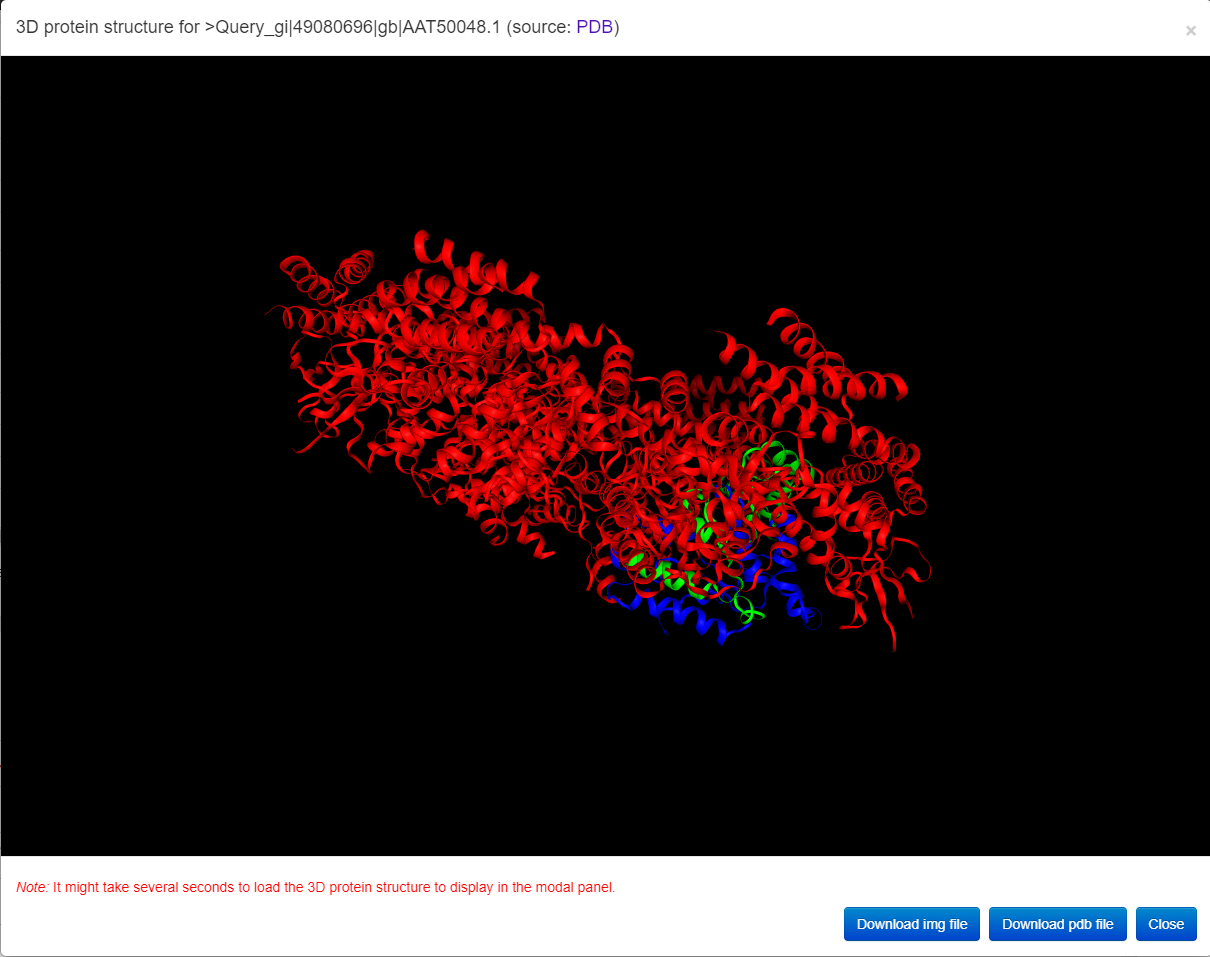
3D structure visualization: To get the protein 3D structure, we use two steps: 1) we make a request to PDB to get a real 3D structure; 2) if there is no hit, we make a request to AlphaFold DB to get a predicted 3D structure. The AlphaFold DB now covers all 'reviewed' proteins in the UniProt Database and will cover all proteins in the uniref90 dataset in 2022. Using this way, we aim to provide 3D structure visualizations for the majority of predicted substrates.
Note: Functional analysis results are showed in the result page (described in the following section) if users select any of them.
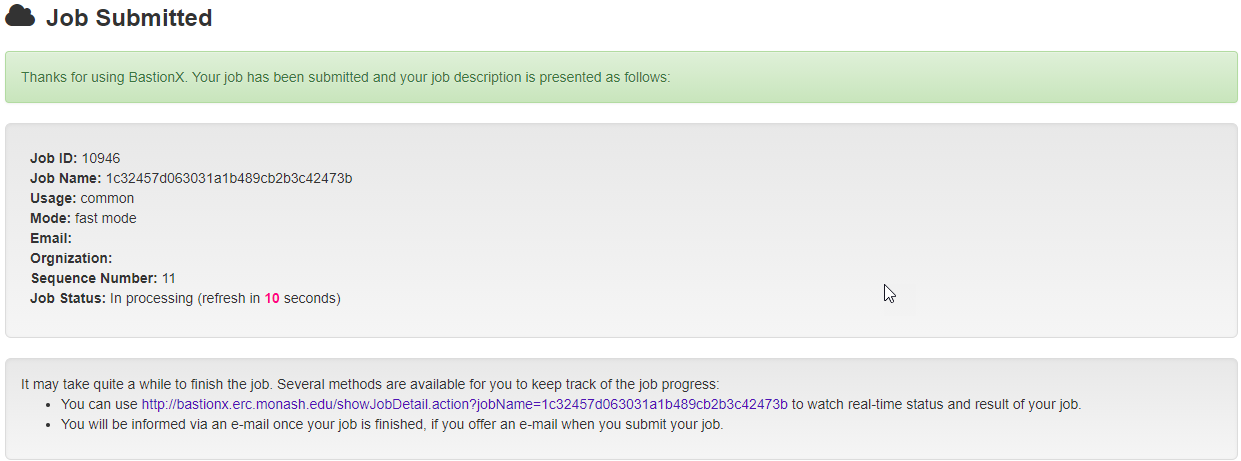
Once a job is submitted, a unique link will be generated to refer to the job summary page during the job execution process (shown in the following figure). Users could use this link to track their job execution progress, and access or download their prediction results once completed. Users will also be notified by email if they provide one in the input page.
3. BastionX Prediction Result Instructions
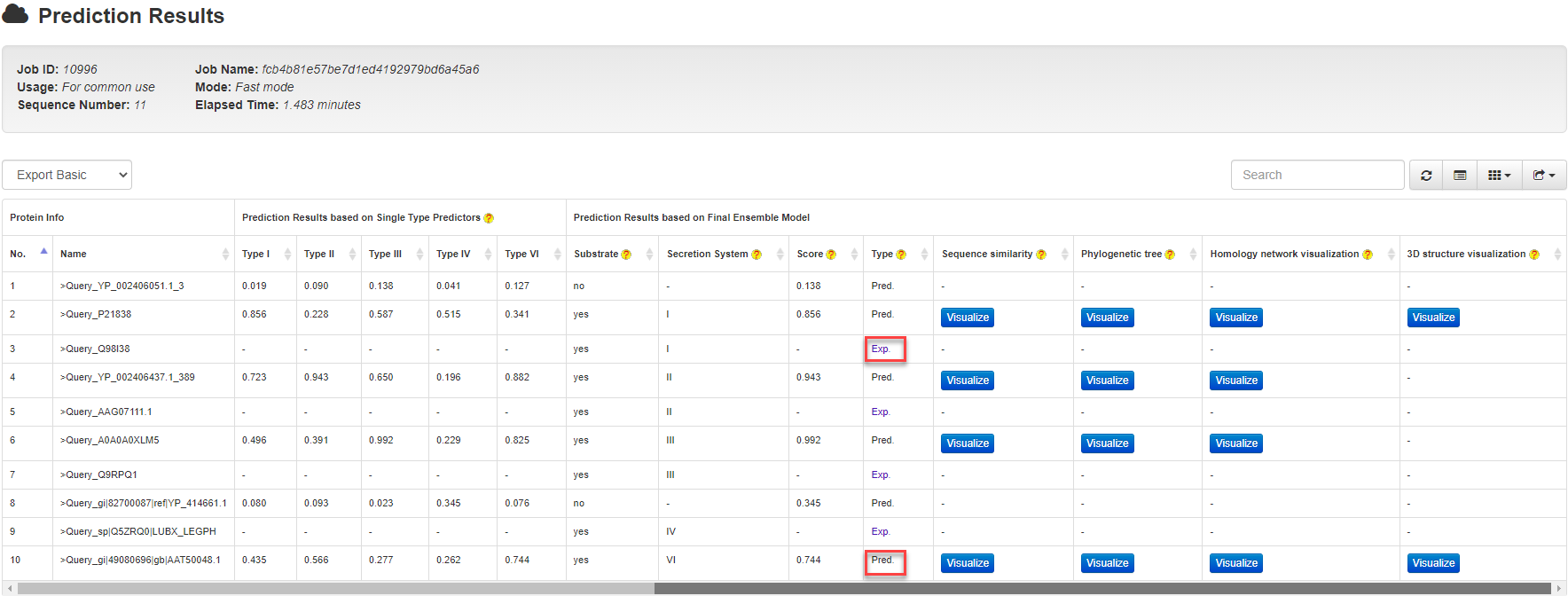
BastionX incorporates a built-in list (continuously updated to keep in pace with BastionHub ) of major types of secreted substrates (including types I, II, III, IV and VI secreted substrates) to annotate the prediction results. This will distinguish known substrates (marked with Exp.) from the computationally predicted ones (marked with Pred.) and additionally provide detailed annotations of those known substrates for users (illustrated in the following figure).
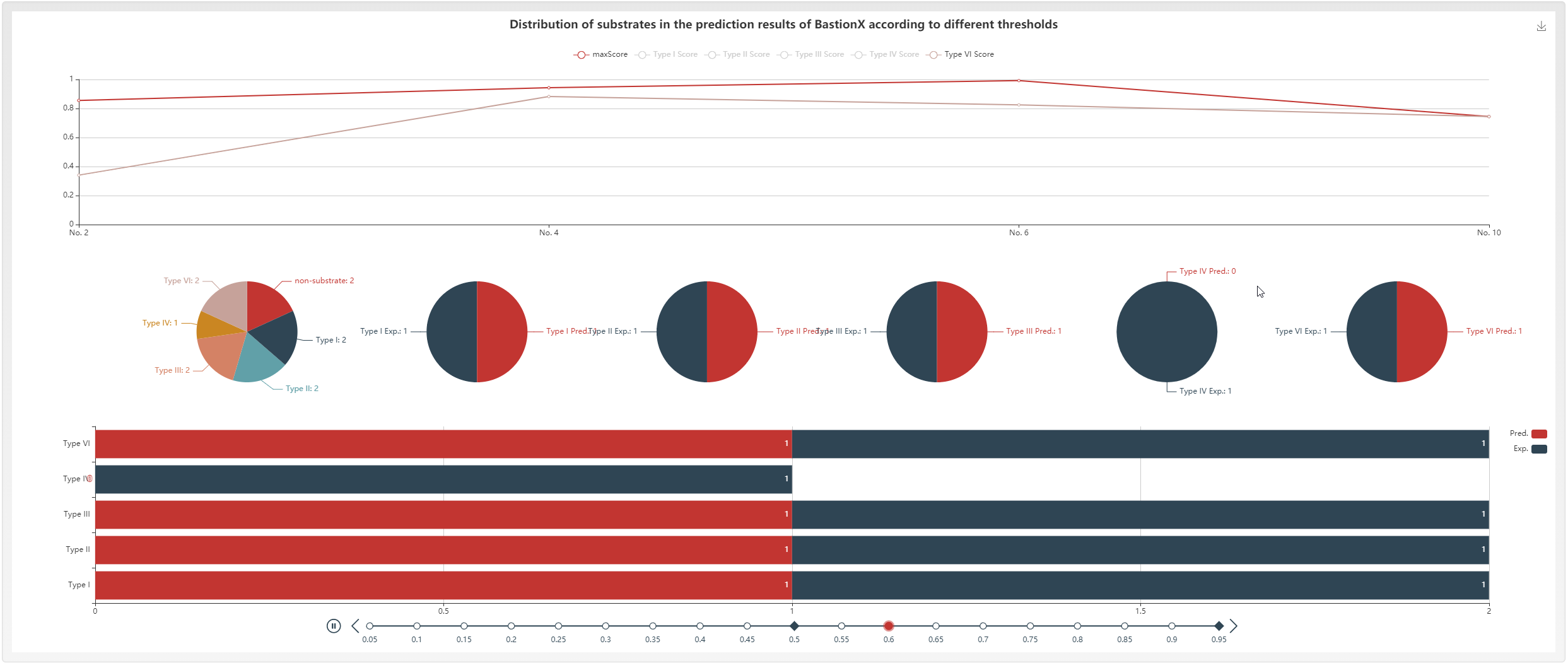
Besides, BastionX provides the interactive visualization to provide a landscape of substrate distributions in query protein sequences.
For a known secreted substrate (e.g. UniProt ID: Q98I38; BastionHub ID: SS02266), the result is marked as Exp. with a URL link to BastionHub to navigate its detailed information .
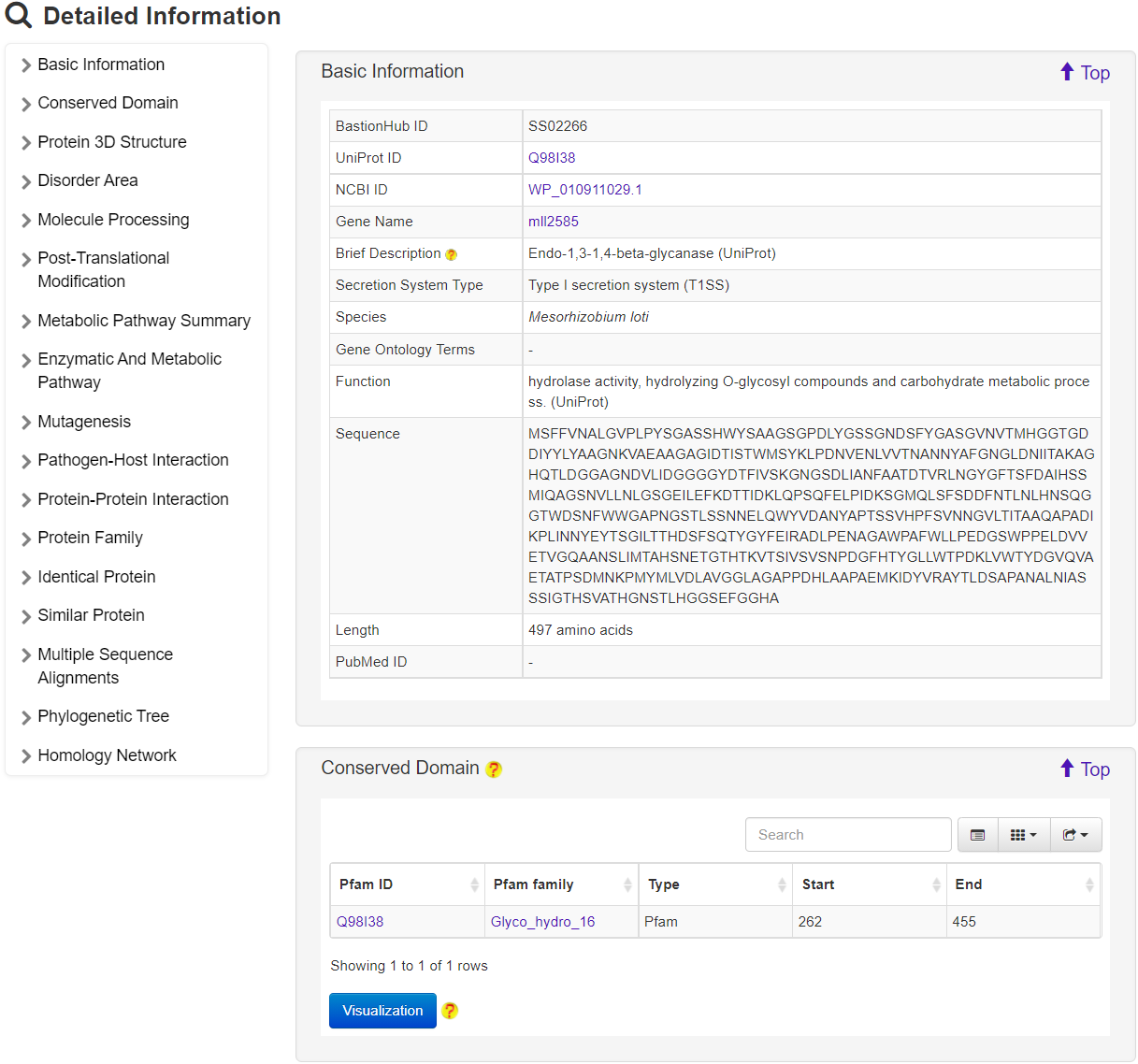
For a computationally predicted secreted substrate, the result is marked as Pred. with detailed prediction results (including those predicted by the single type predictors and the final integrative predictor). If users activate any of four functional analysis options in the input page, the corresponding results for each of predicted secreted substrates will be visualized via clicking the Visualize button.
The result of Similarity analysis is provided in the following figure.
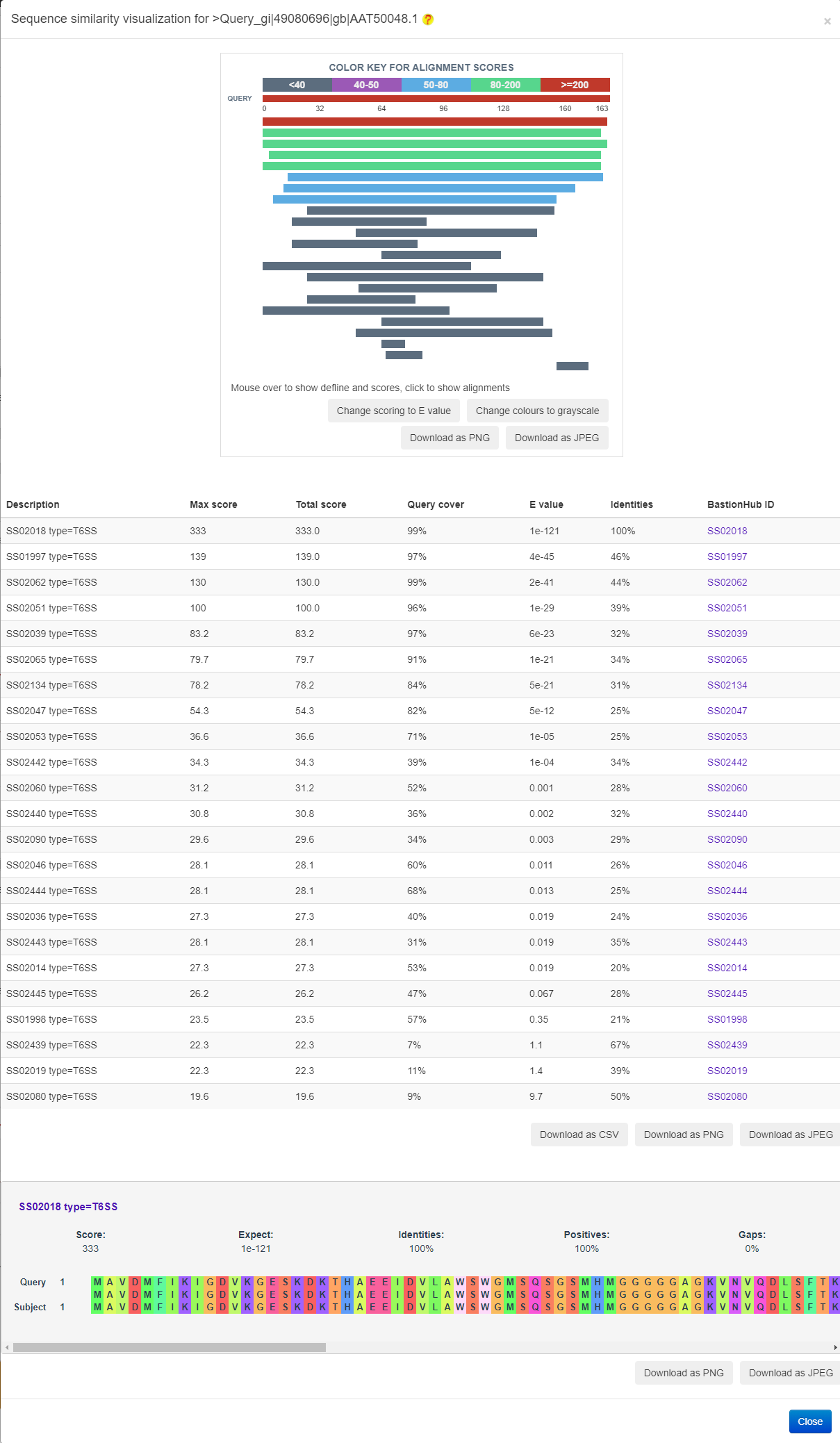
The result of Phylogenetic analysis is provided in the following figure.
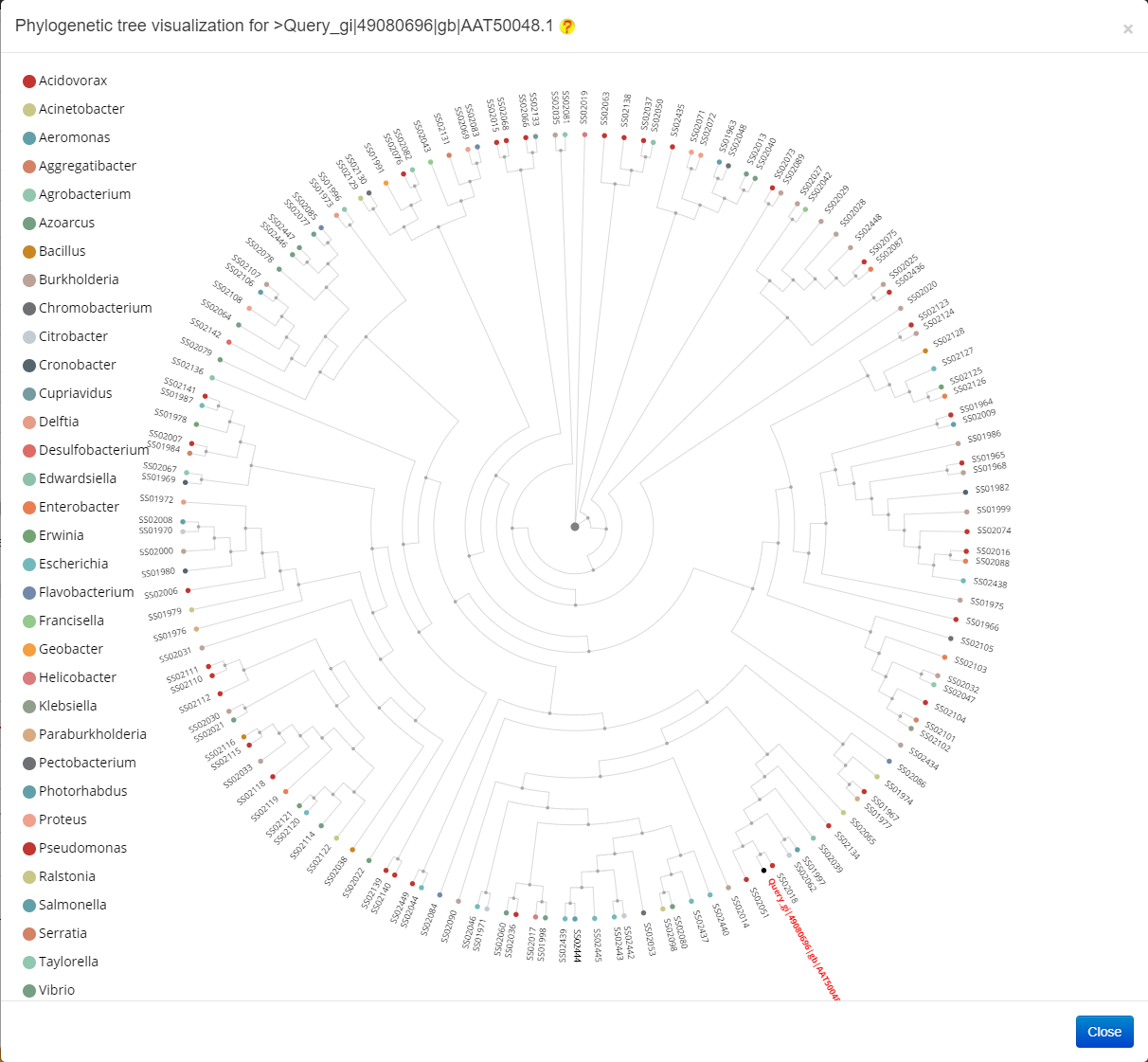
The result of Homology network analysis is provided in the following figure.
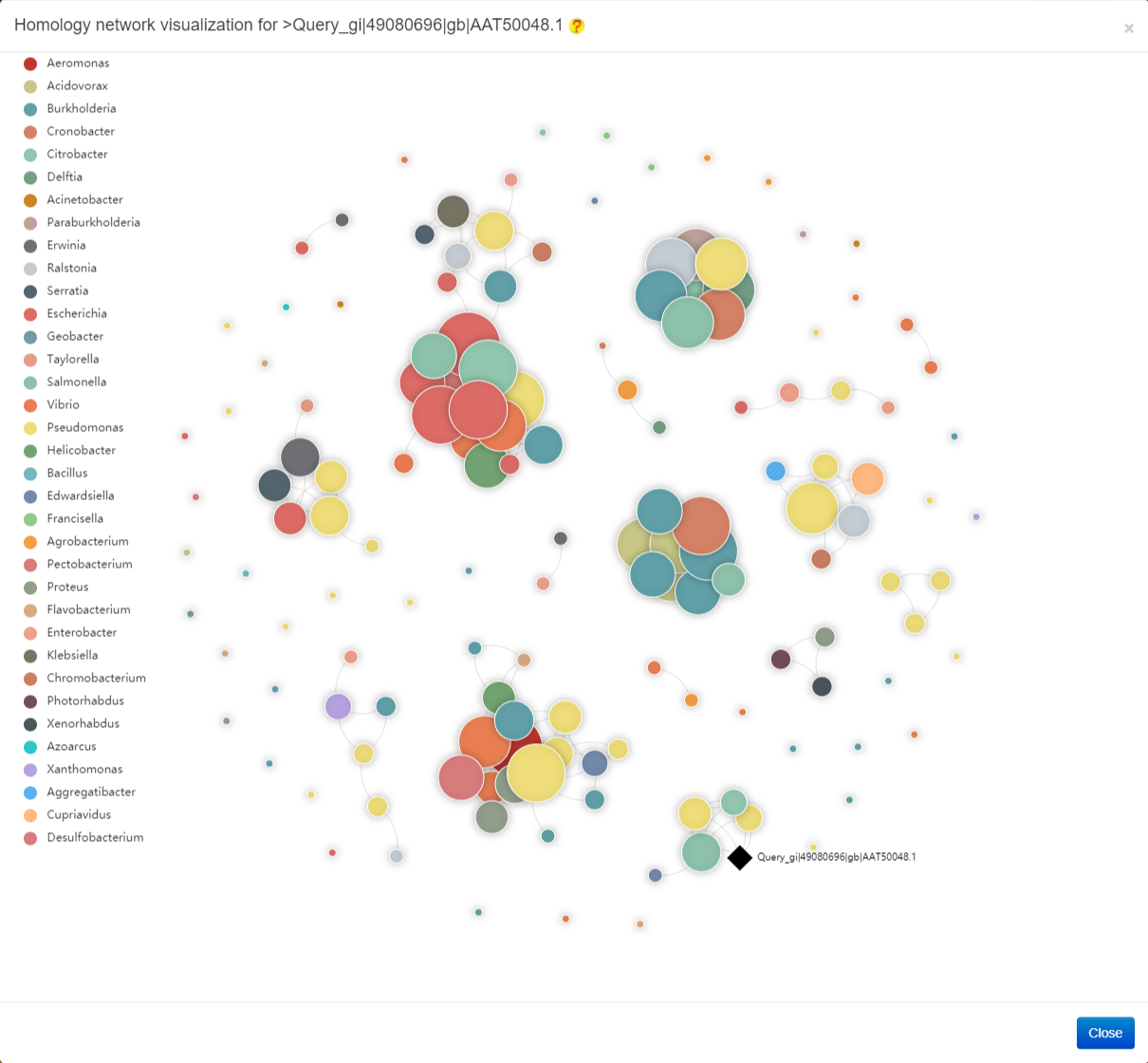
The interactive 3D protein structure is provided in the following figure. Users can click the buttons in the bottom right corner to download the image or original pdb (Protein Data Bank) file.
STANDALONE SOFTWARE
1. Overview
The standalone toolkit of BastionX (open source) can be downloaded at the DOWNLOAD page.
2. Using BastionX
For users who have the capacity to perform high-throughput generation of intermediate files (i.e. using the blast programme to generate .pssm files against uniref 50/90/100 databases) for a very large dataset using their local computers, an open source standalone toolkit was also developed to meet their demands. The standalone version of BastionX was developed using Perl, Python and R, and can be executed on Unix/Linux, Windows and Mac OS. As an open source software, it allows users to freely access, modify and redistribute its source codes, which enables users to tailor BastionX according to their specific requirements.
2.1 System Requirements
-
Operating systems:
Windows,Unix/Linux,Mac OS - Dependencies:
2.2 File Description in the Toolkit Directory
-
input: The input file folder (users can specify their own input file folder using-i).pssm_files: The PSSM file folder (users can specify their own PSSM file folder using-p), which contains the example PSSM files.example_1.pssm,example_2.pssm: The example PSSM files.example.fasta: The example fasta file used to predict substrates.
-
output: The folder used to store the BastionX prediction results (users can specify their own output folder using-o).*.csvfile (example_result.csv): The prediction result file of the example fasta fileexample.fasta.
-
utils: The folder used to store a bunch of utility scripts that aiding users to formalize fasta sequences and predict substrates.removeIllegalSequences.pl: A Perl 5 script used to remove fasta sequences containing illegal characters, such as 'B', 'J', 'O', 'U', 'X' and 'Z'.
# usage examples: perl removeIllegalSequences.pl -i example.fasta -o example_corrected.fasta
removeShortSequences.pl: A Perl 5 script used to remove fasta sequences shorter than a given threshold value (-n).
# usage examples: perl removeShortSequences.pl -i example.fasta -o example_corrected.fasta -n 50 perl removeShortSequences.pl -i example.fasta -o example_corrected.fasta -n 100
DIFFUSER_Standalone_Toolkit : A toolkit to generate machine learning features based on protein, DNA and RNA sequences.txss_multiple_read_model_predict_vote.R : A R script used to predict different types of substrates based on generated features.models: The folder used to store machine learning based models.
tmp: The folder used to cache temporary files in the execution process of the programme.
docs: The folder used to store instruction documents.
userguide.pdf: The detailed user manual for the BastionX standalone toolkit.
bastionx_standalone.pl: The Perl 5 based entry programme to invoke and run the BastionX standalone toolkit.
2.3 Usage
Data preparation:
Two types of input files are required for BastionX:
-
fasta file: A fasta file should contain one/multiple protein sequences in fasta format. Users can specify a fasta file as input by using-iparameter. -
pssm_files: PSSM files for the fasta file (using BLAST against uniref 50/90/100 databases) should be provided in a certain folder, which should be specified by users using-pparameter.
Command line examples: For Unix/Linux/Mac OS X users:
perl bastionx_standalone.pl -i input/example.fasta -o output/example_result.csv -p input/pssm_files -m fast
For Windows users:
perl bastionx_standalone.pl -i input/example.fasta -o output/example_result.csv -p input/pssm_files -m fastOr
perl bastionx_standalone.pl -i input\example.fasta -o output\example_result.csv -p input\pssm_files -m accurateOr
perl bastionx_standalone.pl -i input\\example.fasta -o output\\example_result.csv -p input\\pssm_files -m balanced
NOTE: The main usage difference between Windows and other OS is the file path format. BastionX allows /,\,\\ as path separators on windows in accordance with users' habits.
Parameters:
-
-i: Specify the input file path of a file in fasta format. -
-o: Specify the output file path for the prediction results (in.csvor.txt). -
-p: Specify the PSSM file folder path. -
-m: Specify any of the interested modes includingfast,accurateandbalanced.
For -i,-o and -p, absolute and relative paths are both allowed.
2.4 Input File Check
For the input file in fasta format, if there exsits sequence(s) shorter than 30 or containing illegal characters, such as B, J, O, U, X and Z, the program will exit and show corresponding tips.
Please refer to the output message, accordingly use the utility scripts in utils folder to dispose of the fasta sequences, and then try again.
2.5 Annotation of the Prediction Results:
-
Prediction results are represented in
.csvor.txtformat.